Support:Documents:Examples:Model Corrected Input Function
Model-Corrected Input Function
Overview
For 18F-FDG PET studies, the estimation of kinetic rate constants required knowledge of the input function (i.e., 18F-FDG plasma time activity curve). The standard method to determine the input function is to measure 18F-FDG activity concentration in the arterial blood. However, this invasive procedure is limited by the small size of blood vessels and the limited blood volume. One alternative way is to estimate the input function non-invasively. One of non-invasive methods is image-derived input functions (IDIFs), which estimate input function from a region of interest (ROI) within the ventricular cavity. However, due to the limited spatial resolution and the cardiac and respiratory motion, cross-contamination from surrounding tissues to vascular structures affects the accuracy of estimated input function. To solve these problems, Fang proposed a model-corrected input function (MCIF), which used simultaneous estimation to correct the spillover and partial volume effect for IDIFs.
Example of Estimating Input Function Using MCIF
Here is an example of COMKAT GUI for estimating kinetic rate constants, and the MCIF method is used to estimate input function.
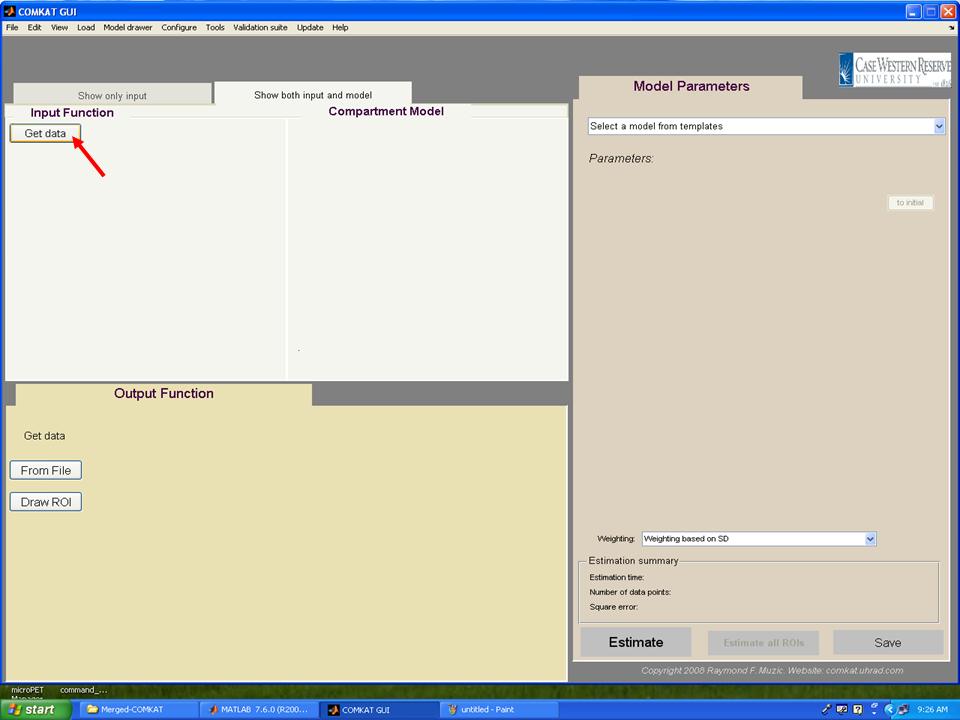
Step 1. Type "comkat" in Matlab commandwindow and "COMKAT_GUI" GUI (below figure) should be started.
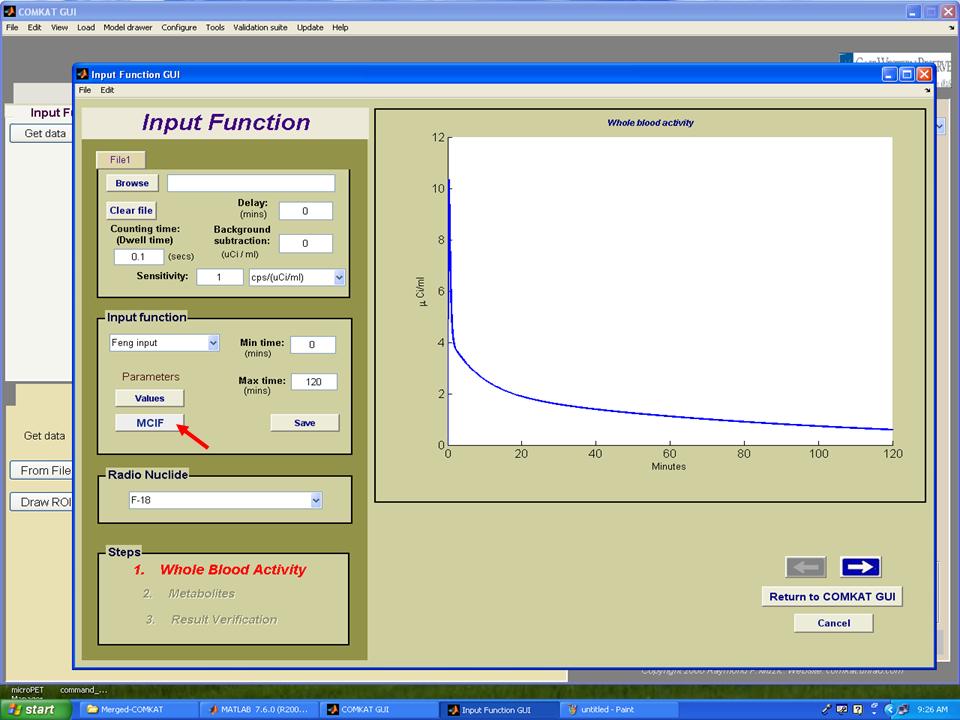
Step 2. Hit 'Get data' button in "COMKAT_GUI" and "Input Function GUI" (below figure) shoud be started.
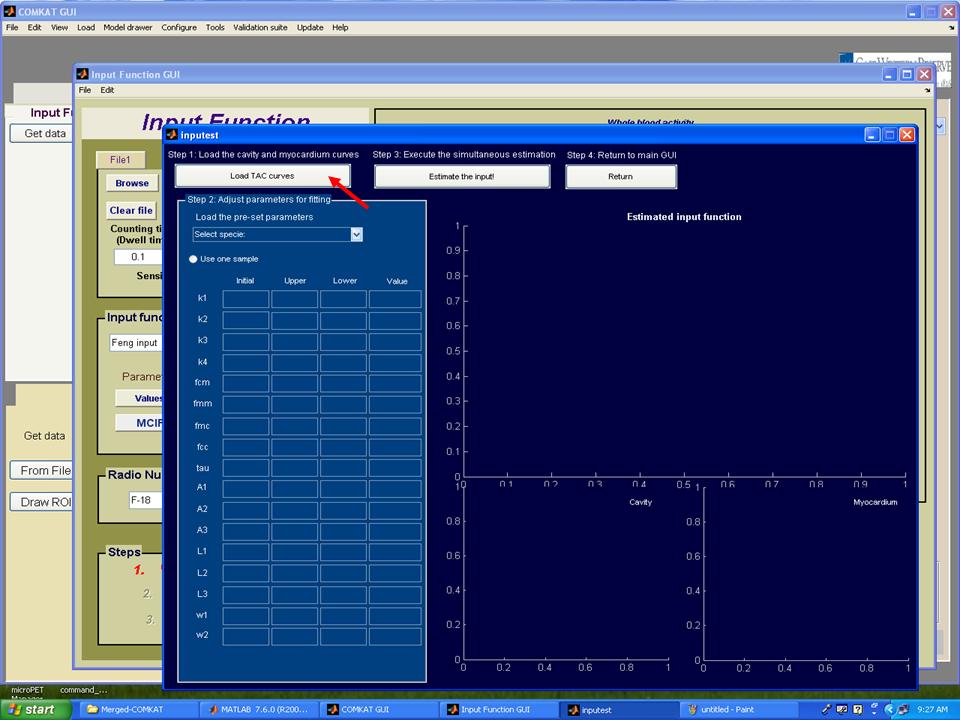
Step 3. Hit 'MCIF' button in "Input Function GUI" and "inputest" GUI (below figure) shoud be started.
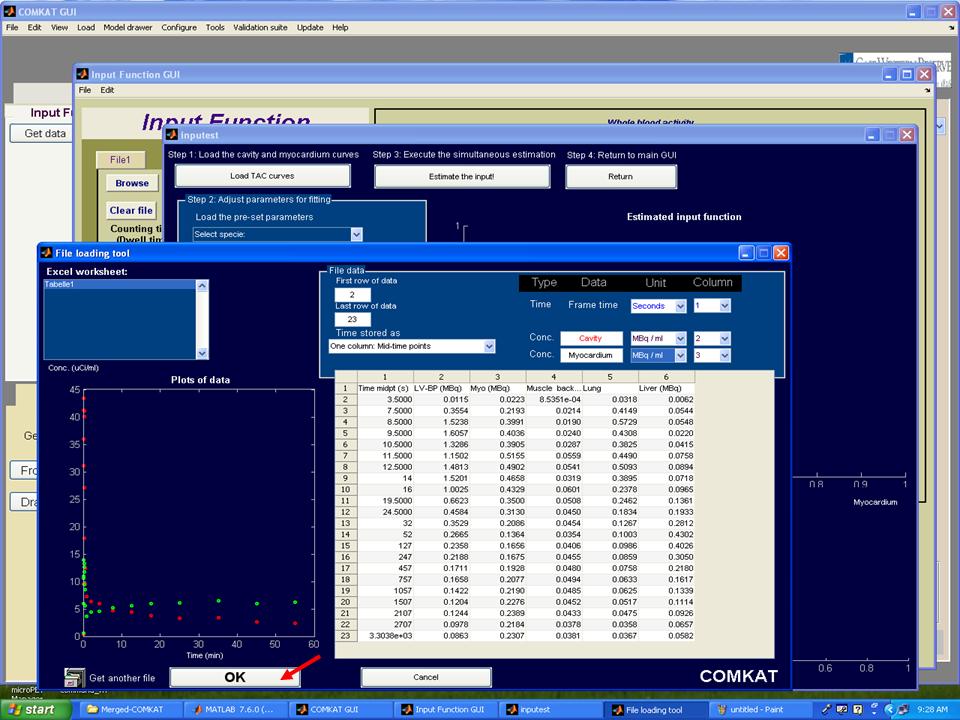
Step 4. Hit 'Load TAC curves' button in "inputest" GUI and "File loading tool" GUI (below figure) shoud be started. Then, choose concentrations of cavity and myocardium from the file user loaded and set units that user defined. Hit 'OK' button to return to "inputest" GUI.
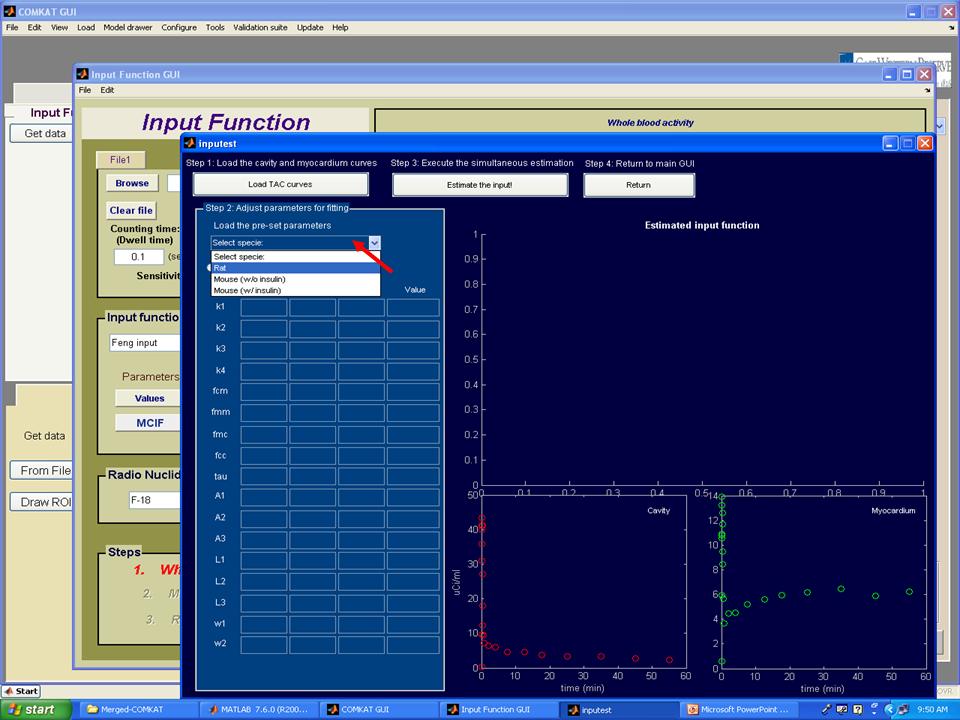
Step 5. Now, the TAC curves for cavity and myocardium should appear in "inputest" GUI. Then, hit 'Load the pre-set parameters' to choose the animal model you used. Note: pre-set parameters may change with different studies and user should try different initial, upper and lower values.
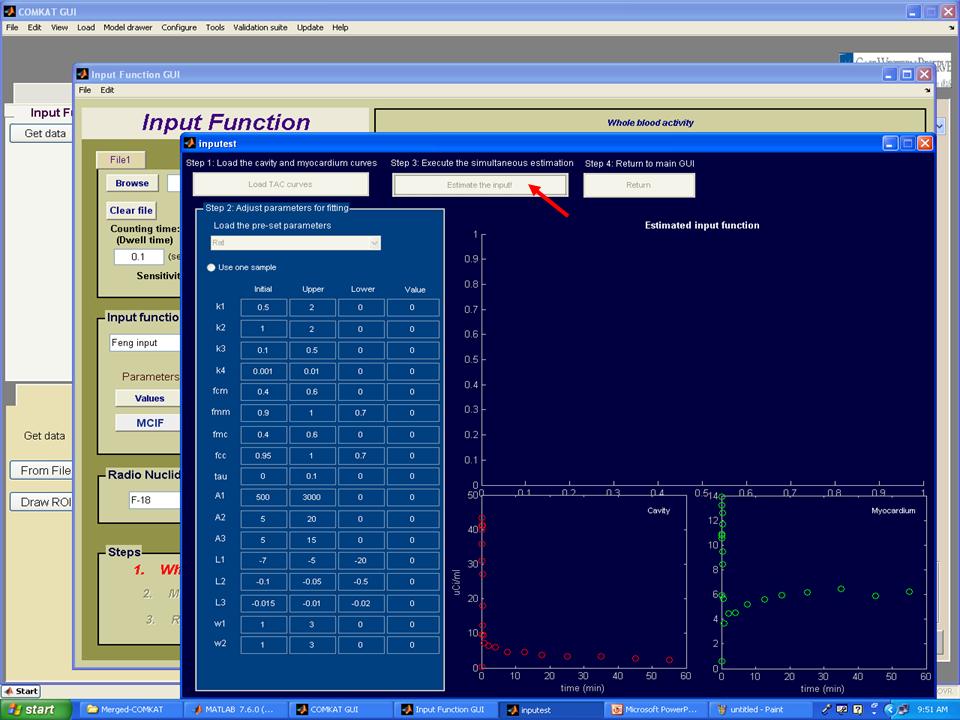
Step 6. Hit 'Estimate the input' button to start the estimation of parameters. Note: please wait several minutes until the estimation process completed.
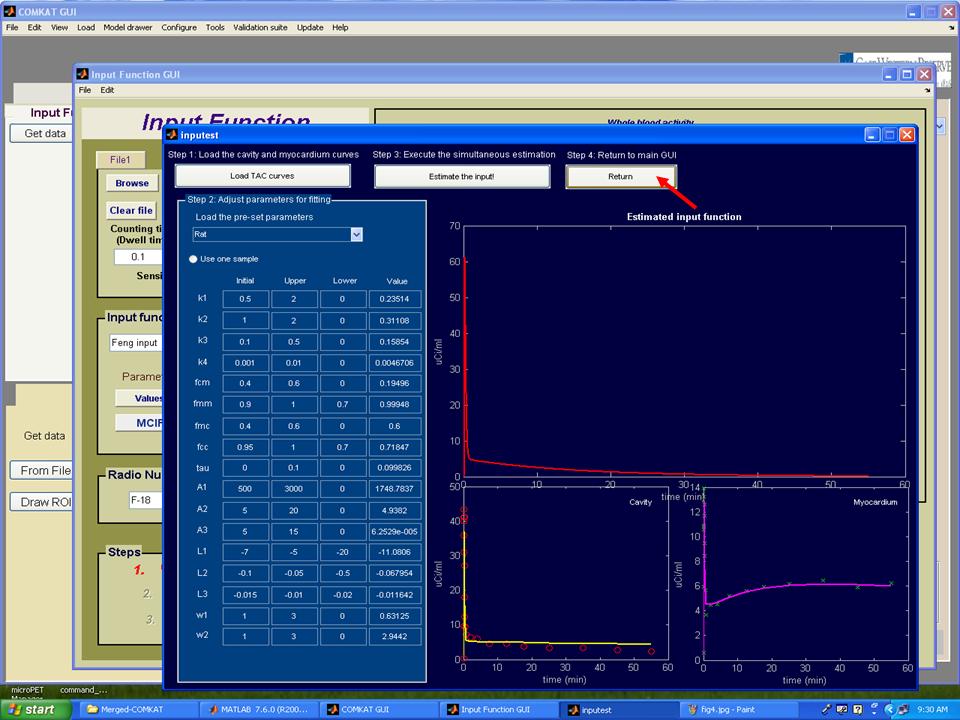
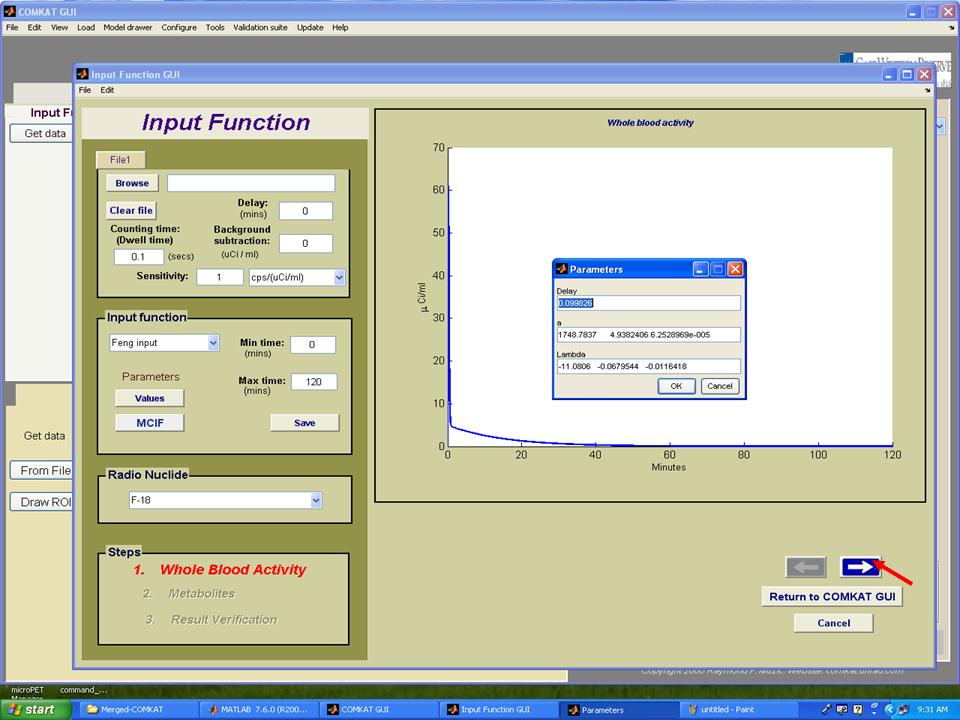
Step 7. As the estimation process completed, the TAC curve for estimated input function should appear in "inputest" GUI and the estimated parameters should in the column 'Value'. Then, hit 'Return' button to return to "Input Function GUI" (below figure) and the TAC curve for whole blood activity should appear. Note: hit 'Values' to see estimated parameters that define the input function. Hit the arrow to the next step.
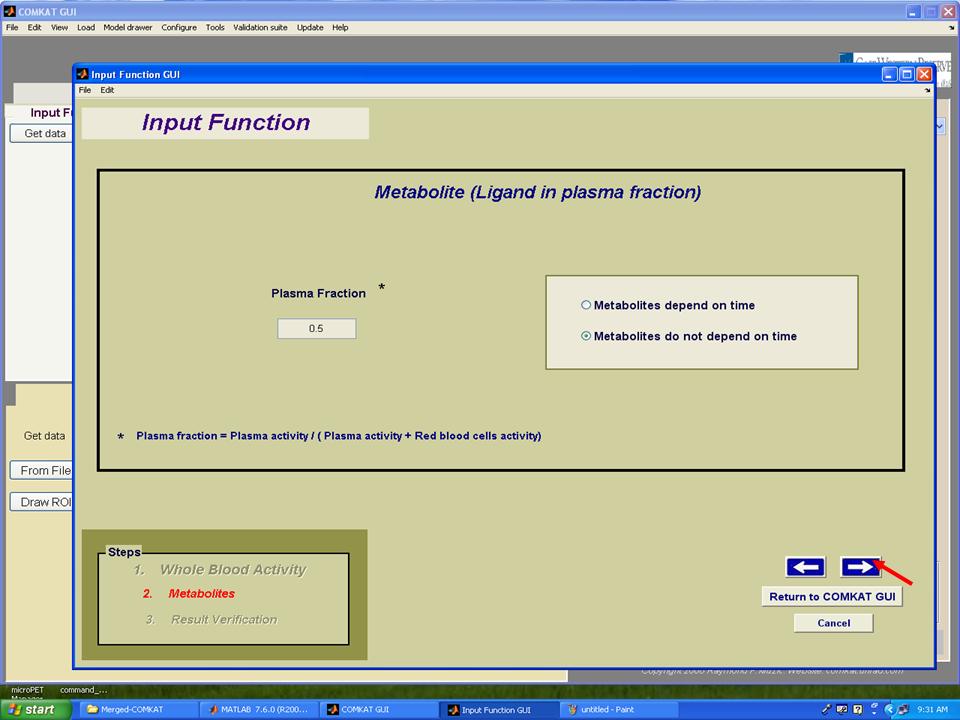
Step 8. Set plasma fraction and then hit the arrow to the next step.
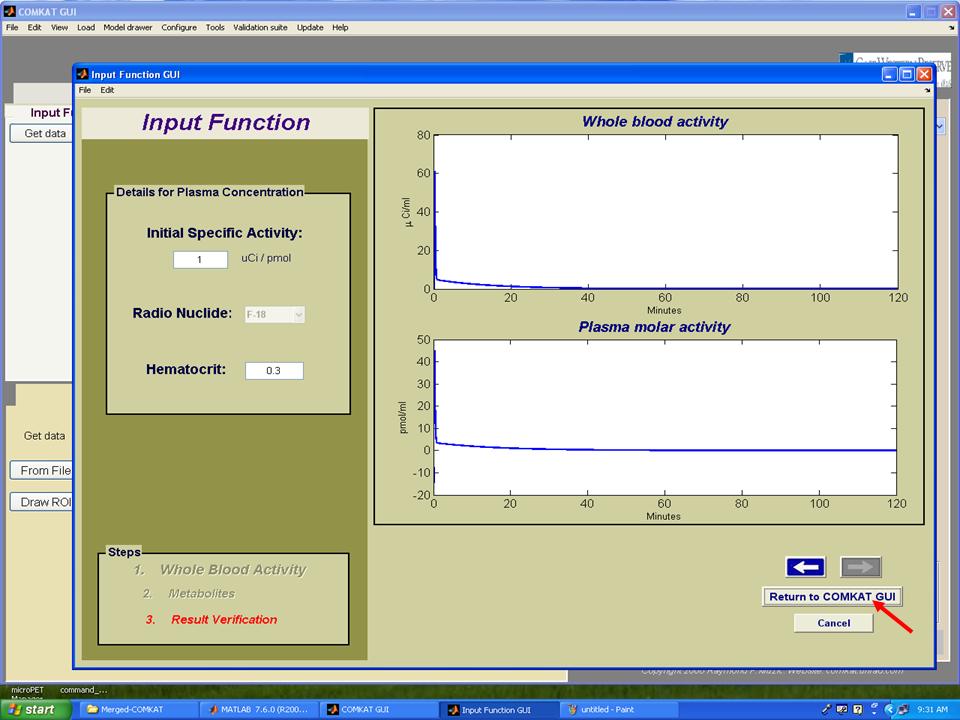
Step 9. Set initial specific activity and hematocrit for producing TAC curve for plasma and then hit 'Return to COMKAT GUI' to return to "COMKAT GUI".
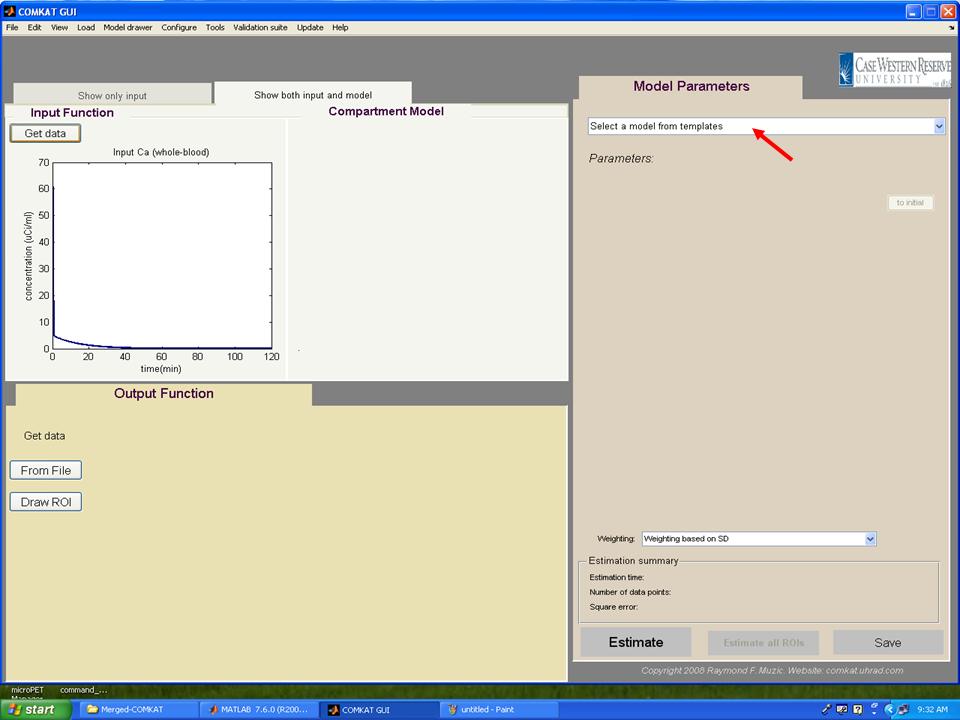
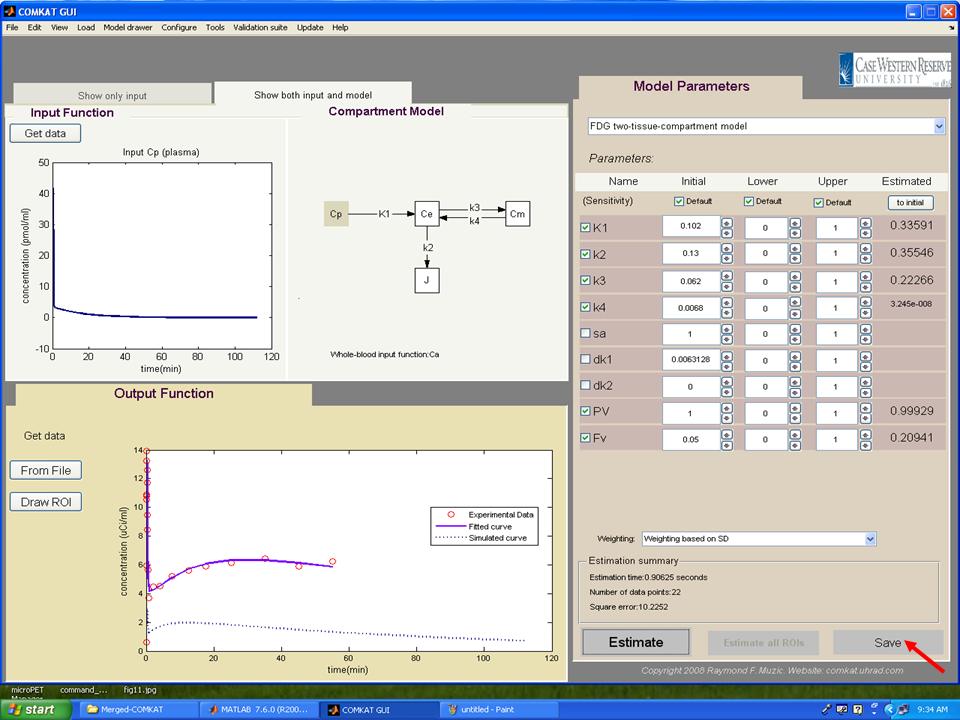
Step 10. The TAC curve for whole bood should appear in "COMKAT GUI" (below figure). Then, hit 'Select a model from templates' to choose the kinetic model.
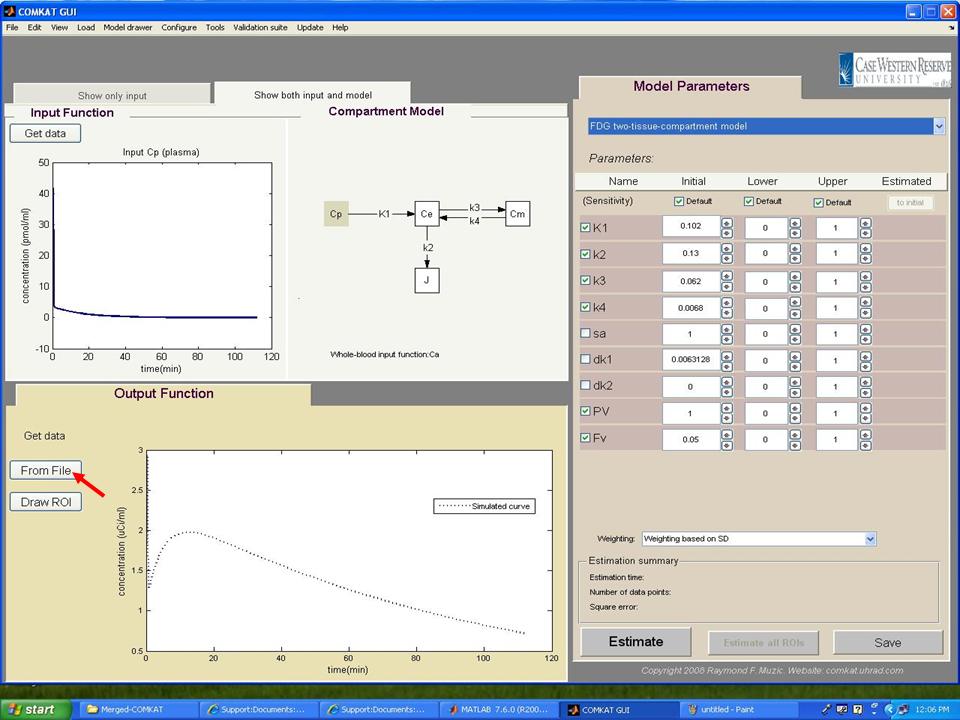
Step 11. In "COMKAT GUI" (below figure), the initial, upper and lower kinetic rate constants for the model should appear in the right side and