Difference between revisions of "Support:Documents:Examples:Estimate Input Delay and Rate Constants"
(New page: == Estimating Input Delay and Rate Constants == This example demonstrates an approach to simultaneously estimating input function delay along with parameters of the 1-tissue compartment (e...) |
|||
| Line 1: | Line 1: | ||
== Estimating Input Delay and Rate Constants == | == Estimating Input Delay and Rate Constants == | ||
This example demonstrates an approach to simultaneously estimating input function delay along with parameters of the 1-tissue compartment (e.g. blood flow) model. | This example demonstrates an approach to simultaneously estimating input function delay along with parameters of the 1-tissue compartment (e.g. blood flow) model. | ||
| − | For the sake of generality, | + | For the sake of generality, this could be interpreted as a |
| − | |||
| Line 48: | Line 47: | ||
ppdMdtau = mkpp(breaks, repmat(k-1:-1:1,d*l,1) .* coefs(:,1:k-1), d); | ppdMdtau = mkpp(breaks, repmat(k-1:-1:1,d*l,1) .* coefs(:,1:k-1), d); | ||
</pre> | </pre> | ||
| − | |||
| − | |||
| − | |||
The input function is then implemented as | The input function is then implemented as | ||
| Line 72: | Line 68: | ||
| + | ==== Pixel values ==== | ||
| + | In this example, the pixel values are assumed to represent measurements of C<sub>T</sub>. (To keep the example simple, intravascular concentration of tracer is ignored.) | ||
| + | The values of the parameters p = [K<sub>1</sub>; k<sub>2</sub>; tau] will be estimated. | ||
| + | |||
| + | |||
| + | == Example: Implement the Compartment Model == | ||
| + | <pre> | ||
| + | % create new, empty model | ||
| + | cm = compartmentModel; | ||
| + | |||
| + | % define default values for parameters | ||
| + | cm = addParameter(cm, 'k1', 0.3); | ||
| + | cm = addParameter(cm, 'k2', 0.5); | ||
| + | cm = addParameter(cm, 'tau', 0.25); | ||
| + | cm = addParameter(cm, 'sa', 1); % specific activity at t=0 | ||
| + | cm = addParameter(cm, 'dk', 0.34); % decay constant | ||
| + | cm = addParameter(cm, 'tau', 0.25); % time delay | ||
| + | % add compartments | ||
| + | cm = addCompartment(cm, 'CT'); | ||
| + | cm = addCompartment(cm, 'J'); | ||
| + | |||
| + | % define the input(first column is time, second it concentration) | ||
| + | inputData = [ | ||
| + | 0 0 | ||
| + | 0.0500 0 | ||
| + | 0.1000 0 | ||
| + | 0.1500 14.5234 | ||
| + | 0.2000 52.1622 | ||
| + | 0.2500 76.6730 | ||
| + | 0.3000 91.6416 | ||
| + | 0.4000 103.0927 | ||
| + | 0.5000 100.9401 | ||
| + | 0.7000 83.5343 | ||
| + | 0.8000 74.3628 | ||
| + | 1.0000 59.7726 | ||
| + | 1.2500 48.7530 | ||
| + | 1.5000 43.0772 | ||
| + | 1.7500 40.1924 | ||
| + | 2.0000 38.6236 | ||
| + | ]; | ||
| + | |||
| + | % determine spline coefficients for linear interpolation | ||
| + | ppM = lspline(inputData(:,1), inputData(:,2)); | ||
| + | % analytically calculate derivative | ||
| + | [breaks, coefs, l, k, d] = unmkpp(ppM); | ||
| + | ppdMdtau = mkpp(breaks, repmat(k-1:-1:1,d*l,1) .* coefs(:,1:k-1), d); | ||
| + | X = {ppM, ppdMdtau}; | ||
| + | |||
| + | cm = addInput(cm, 'Cp','sa','dk', 'DelayExampleInput', 'tau', X); | ||
| + | |||
| + | % connect compartments and inputs | ||
| + | cm = addLink(cm, 'L', 'Cp', 'CT', 'k1'); | ||
| + | cm = addLink(cm, 'K', 'CT', 'J', 'k2'); | ||
| + | % define the activity concentration in tissue pixel | ||
| + | cm = addOutput(cm, 'TissuePixel', {'CT','1'}, {'Cp','0'}); | ||
| + | % specify scan frame times | ||
| + | st = [[0:5:85]' [5:5:90]']/60; % division by 60 converts sec to min | ||
| + | cm = set(cm, 'ScanTime', st); | ||
| − | + | % solve the model | |
| − | + | [sol,solIndex] = solve(cm); | |
| − | |||
| + | midScanTime = (st(:,1) + st(:,2)) / 2; | ||
| − | == | + | % Initial conditions and boundary for K1, k2, tau |
| + | pinit=[0.01;0.01;0.01]; | ||
| + | lb =[0.01;0.01;0.01]; | ||
| + | ub =[1;1;5]; | ||
| − | = | + | cm=addSensitivity(cm,'k1','k2','tau'); |
| − | |||
| + | cm=set(cm,'ExperimentalData',sol(:,3)); | ||
| + | [pp,qfitnull,modelfit,pp1,output]=fitGen(cm,pinit,lb,ub,'OLS'); | ||
| − | == | + | % solve model using estimated parameters |
| + | cm1=cm; | ||
| + | cm1=set(cm1,'ParameterValue','k1',pp(1)); | ||
| + | cm1=set(cm1,'ParameterValue','k2',pp(2)); | ||
| + | cm1=set(cm1,'ParameterValue','tau',pp(3)); | ||
| + | est_sol=solve(cm1); | ||
| − | + | plot(midScanTime,sol(:,3),'o',midScanTime,est_sol(:,3),'r-'); | |
| + | xlabel('Minutes'); | ||
| + | ylabel('KBq/ml'); | ||
| + | legend('Data', 'Fit'); | ||
| + | </pre> | ||
Revision as of 21:51, 18 December 2008
Estimating Input Delay and Rate Constants
This example demonstrates an approach to simultaneously estimating input function delay along with parameters of the 1-tissue compartment (e.g. blood flow) model. For the sake of generality, this could be interpreted as a
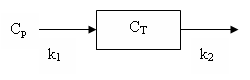
Tissue uptake
This model has one tissue compartment. Material in the blood is assumed to rapidly exchange with that in (extravascular) tissue. The tissue model has two rate constants: K1 and k2 and is depicted in the diagram:
It can also be described by the differential equation:
dCT/dt = K1 Cp - k2 CT
where CT is the tissue concentration and Cp is the plasma concentration of radioactive water.
CT and Cp are interpreted as either molar concentrations (Salinas, Muzic and Saidel 2007).
Input function
This example also demonstrates how to temporally align measured input function data to the tissue (image) data. Here the plasma concentration vs. time t is modeled as
Cp(t, tau) = M(t-d) if t >= tau; 0 if t < tau .
M(t) is the measured input data, tau is the delay parameter to estimate, and t is the time in minutes.
Because the delay tau is estimated in this example, it will be handy to note that derivative of the input with respect to delay is
Cp(t)/d tau = - dM/dt|t-tau
For this example, M is determined by using linear interpolation to interpolate between measured values. The interpolation is implemented using piecewise polynomial framework (and it could be easily modified to accommodate cubic spline interpolation).
Let inputData be a two-column matrix with column 1 holding sample times and column 2 holding the sampled input concentrations.
The interpolation coefficients may be calculated using the lspline function
ppM = lspline(inputData(:,1), inputData(:,2));
To evaluate the derivative dCp/dt, which can be determined in terms of dM/dt by analytically differentiating the interpolating polynomial.
[breaks, coefs, l, k, d] = unmkpp(ppM); ppdMdtau = mkpp(breaks, repmat(k-1:-1:1,d*l,1) .* coefs(:,1:k-1), d);
The input function is then implemented as
function [Cp, dCpdtau] = DelayExampleInput(parm, t, X)
if (nargout > 0),
t = t(:); % ensure t is a column vector
tau = parm(1); % delay
ppC = X{1}; % piecewise-polynomial coefficients for Cp
Cp = zeros(size(t));
f = find(t > tau);
Cp(f) = ppval(ppC, t(f) - tau);
if (nargout > 1),
ppdCdt = X{2}; % piecewise-polynomial coefficients for derivative
dCpdtau = zeros(size(t));
dCpdtau(f) = -ppval(ppdCdt, t(f) - tau);
end
end
Pixel values
In this example, the pixel values are assumed to represent measurements of CT. (To keep the example simple, intravascular concentration of tracer is ignored.) The values of the parameters p = [K1; k2; tau] will be estimated.
Example: Implement the Compartment Model
% create new, empty model
cm = compartmentModel;
% define default values for parameters
cm = addParameter(cm, 'k1', 0.3);
cm = addParameter(cm, 'k2', 0.5);
cm = addParameter(cm, 'tau', 0.25);
cm = addParameter(cm, 'sa', 1); % specific activity at t=0
cm = addParameter(cm, 'dk', 0.34); % decay constant
cm = addParameter(cm, 'tau', 0.25); % time delay
% add compartments
cm = addCompartment(cm, 'CT');
cm = addCompartment(cm, 'J');
% define the input(first column is time, second it concentration)
inputData = [
0 0
0.0500 0
0.1000 0
0.1500 14.5234
0.2000 52.1622
0.2500 76.6730
0.3000 91.6416
0.4000 103.0927
0.5000 100.9401
0.7000 83.5343
0.8000 74.3628
1.0000 59.7726
1.2500 48.7530
1.5000 43.0772
1.7500 40.1924
2.0000 38.6236
];
% determine spline coefficients for linear interpolation
ppM = lspline(inputData(:,1), inputData(:,2));
% analytically calculate derivative
[breaks, coefs, l, k, d] = unmkpp(ppM);
ppdMdtau = mkpp(breaks, repmat(k-1:-1:1,d*l,1) .* coefs(:,1:k-1), d);
X = {ppM, ppdMdtau};
cm = addInput(cm, 'Cp','sa','dk', 'DelayExampleInput', 'tau', X);
% connect compartments and inputs
cm = addLink(cm, 'L', 'Cp', 'CT', 'k1');
cm = addLink(cm, 'K', 'CT', 'J', 'k2');
% define the activity concentration in tissue pixel
cm = addOutput(cm, 'TissuePixel', {'CT','1'}, {'Cp','0'});
% specify scan frame times
st = [[0:5:85]' [5:5:90]']/60; % division by 60 converts sec to min
cm = set(cm, 'ScanTime', st);
% solve the model
[sol,solIndex] = solve(cm);
midScanTime = (st(:,1) + st(:,2)) / 2;
% Initial conditions and boundary for K1, k2, tau
pinit=[0.01;0.01;0.01];
lb =[0.01;0.01;0.01];
ub =[1;1;5];
cm=addSensitivity(cm,'k1','k2','tau');
cm=set(cm,'ExperimentalData',sol(:,3));
[pp,qfitnull,modelfit,pp1,output]=fitGen(cm,pinit,lb,ub,'OLS');
% solve model using estimated parameters
cm1=cm;
cm1=set(cm1,'ParameterValue','k1',pp(1));
cm1=set(cm1,'ParameterValue','k2',pp(2));
cm1=set(cm1,'ParameterValue','tau',pp(3));
est_sol=solve(cm1);
plot(midScanTime,sol(:,3),'o',midScanTime,est_sol(:,3),'r-');
xlabel('Minutes');
ylabel('KBq/ml');
legend('Data', 'Fit');